社区微信群开通啦,扫一扫抢先加入社区官方微信群

社区微信群
社区微信群开通啦,扫一扫抢先加入社区官方微信群

社区微信群
前沿
在处理医学图像时,常常会遇到以Dicom格式保存的医学图像,如CT、MRI等。Dicom文件是需要专门的软件或者通过编程,应用相应的库进行处理。为了能够更好地服务下游任务,例如分割或检测腹腔CT图像中某个病灶组织,需要先将Dicom图像进行读取,脱敏,调窗等步骤,以便于后续的编辑。本文使用python对医学Dicom文件进行相应的处理,相比于封装好的软件,笔者认为自己动手的可操作性更强。
目录
1 导入相应的包
2 读取Dicom图像数据
3 设置CT图像的窗宽和窗位
4 获取Dicom图像的tag信息
5 结果保存及可视化
导入相应的包
# load necessary packages
import matplotlib.pyplot as plt
import pydicom.uid
import sys
from PyQt5 import QtGui
import os
import pydicom
import glob
from PIL import *
import matplotlib.pyplot as plt
from pylab import *
from tkinter.filedialog import *
import PIL.Image as Image其核心是使用了python中的pydicom库来处理dicom文件。
读取Dicom图像数据
have_numpy = True
try:
import numpy
except ImportError:
have_numpy = False
raise
sys_is_little_endian = (sys.byteorder == 'little')
NumpySupportedTransferSyntaxes = [
pydicom.uid.ExplicitVRLittleEndian,
pydicom.uid.ImplicitVRLittleEndian,
pydicom.uid.DeflatedExplicitVRLittleEndian,
pydicom.uid.ExplicitVRBigEndian,
]
# 支持"传输"语法
def supports_transfer_syntax(dicom_dataset):
return (dicom_dataset.file_meta.TransferSyntaxUID in
NumpySupportedTransferSyntaxes)
def needs_to_convert_to_RGB(dicom_dataset):
return False
def should_change_PhotometricInterpretation_to_RGB(dicom_dataset):
return False
# 加载 Dicom图像
def get_pixeldata(dicom_dataset):
"""If NumPy is available, return an ndarray of the Pixel Data.
Raises
------
TypeError
If there is no Pixel Data or not a supported data type.
ImportError
If NumPy isn't found
NotImplementedError
if the transfer syntax is not supported
AttributeError
if the decoded amount of data does not match the expected amount
Returns
-------
numpy.ndarray
The contents of the Pixel Data element (7FE0,0010) as an ndarray.
"""
if (dicom_dataset.file_meta.TransferSyntaxUID not in
NumpySupportedTransferSyntaxes):
raise NotImplementedError("Pixel Data is compressed in a "
"format pydicom does not yet handle. "
"Cannot return array. Pydicom might "
"be able to convert the pixel data "
"using GDCM if it is installed.")
if not have_numpy:
msg = ("The Numpy package is required to use pixel_array, and "
"numpy could not be imported.")
raise ImportError(msg)
if 'PixelData' not in dicom_dataset:
raise TypeError("No pixel data found in this dataset.")
# Make NumPy format code, e.g. "uint16", "int32" etc
# from two pieces of info:
# dicom_dataset.PixelRepresentation -- 0 for unsigned, 1 for signed;
# dicom_dataset.BitsAllocated -- 8, 16, or 32
if dicom_dataset.BitsAllocated == 1:
# single bits are used for representation of binary data
format_str = 'uint8'
elif dicom_dataset.PixelRepresentation == 0:
format_str = 'uint{}'.format(dicom_dataset.BitsAllocated)
elif dicom_dataset.PixelRepresentation == 1:
format_str = 'int{}'.format(dicom_dataset.BitsAllocated)
else:
format_str = 'bad_pixel_representation'
try:
numpy_dtype = numpy.dtype(format_str)
except TypeError:
msg = ("Data type not understood by NumPy: "
"format='{}', PixelRepresentation={}, "
"BitsAllocated={}".format(
format_str,
dicom_dataset.PixelRepresentation,
dicom_dataset.BitsAllocated))
raise TypeError(msg)
if dicom_dataset.is_little_endian != sys_is_little_endian:
numpy_dtype = numpy_dtype.newbyteorder('S')
pixel_bytearray = dicom_dataset.PixelData
if dicom_dataset.BitsAllocated == 1:
# if single bits are used for binary representation, a uint8 array
# has to be converted to a binary-valued array (that is 8 times bigger)
try:
pixel_array = numpy.unpackbits(
numpy.frombuffer(pixel_bytearray, dtype='uint8'))
except NotImplementedError:
# PyPy2 does not implement numpy.unpackbits
raise NotImplementedError(
'Cannot handle BitsAllocated == 1 on this platform')
else:
pixel_array = numpy.frombuffer(pixel_bytearray, dtype=numpy_dtype)
length_of_pixel_array = pixel_array.nbytes
expected_length = dicom_dataset.Rows * dicom_dataset.Columns
if ('NumberOfFrames' in dicom_dataset and
dicom_dataset.NumberOfFrames > 1):
expected_length *= dicom_dataset.NumberOfFrames
if ('SamplesPerPixel' in dicom_dataset and
dicom_dataset.SamplesPerPixel > 1):
expected_length *= dicom_dataset.SamplesPerPixel
if dicom_dataset.BitsAllocated > 8:
expected_length *= (dicom_dataset.BitsAllocated // 8)
padded_length = expected_length
if expected_length & 1:
padded_length += 1
if length_of_pixel_array != padded_length:
raise AttributeError(
"Amount of pixel data %d does not "
"match the expected data %d" %
(length_of_pixel_array, padded_length))
if expected_length != padded_length:
pixel_array = pixel_array[:expected_length]
if should_change_PhotometricInterpretation_to_RGB(dicom_dataset):
dicom_dataset.PhotometricInterpretation = "RGB"
if dicom_dataset.Modality.lower().find('ct') >= 0: # CT图像需要得到其CT值图像
pixel_array = pixel_array * dicom_dataset.RescaleSlope + dicom_dataset.RescaleIntercept # 获得图像的CT值
pixel_array = pixel_array.reshape(dicom_dataset.Rows, dicom_dataset.Columns*dicom_dataset.SamplesPerPixel)
return pixel_array, dicom_dataset.Rows, dicom_dataset.Columns
读取到dicom文件中的数据后,实质上是几个图像矩阵,这个过程同时也处理了“脱敏”问题。
设置CT图像的窗宽和窗位
def setDicomWinWidthWinCenter(img_data, winwidth, wincenter, rows, cols):
img_temp = img_data
img_temp.flags.writeable = True
min = (2 * wincenter - winwidth) / 2.0 + 0.5
max = (2 * wincenter + winwidth) / 2.0 + 0.5
dFactor = 255.0 / (max - min)
for i in numpy.arange(rows):
for j in numpy.arange(cols):
img_temp[i, j] = int((img_temp[i, j]-min)*dFactor)
min_index = img_temp < min
img_temp[min_index] = 0
max_index = img_temp > max
img_temp[max_index] = 255
return img_temp该函数的输入变量winwidth和wincenter即为需要设置的窗宽和窗位,这两个值根据研究的问题(不同的组织器官对应不同的窗宽和窗位,有时候也要根据图像效果进行一定的调整)调整不同的值。网上有很多关于相关的窗位和窗宽对应值,这里给出一些参考资料,如果遇到不确定的,最好借鉴查阅相应领域的论文。
Windowing (CT) | Radiology Reference Article | Radiopaedia.orgradiopaedia.org获取Dicom图像的tag信息
def loadFileInformation(filename):
information = {}
ds = pydicom.read_file(filename)
information['PatientID'] = ds.PatientID
information['PatientName'] = ds.PatientName
information['PatientBirthDate'] = ds.PatientBirthDate
information['PatientSex'] = ds.PatientSex
information['StudyID'] = ds.StudyID
information['StudyDate'] = ds.StudyDate
information['StudyTime'] = ds.StudyTime
information['InstitutionName'] = ds.InstitutionName
information['Manufacturer'] = ds.Manufacturer
print(dir(ds))
print(type(information))
return information这个步骤视需求而定,如果不需要查看dicom文件的具体tag信息,此步骤可以跳过。
结果保存及可视化
可以单张保存,或者批量处理。
读取单张dicom文件
def main_single():
dcm = dicom.read_file('81228816') # load dicom_file
# 得到 CT 值,图像的 长, 宽
pixel_array, dcm.Rows, dcm.Columns = get_pixeldata(dcm)
# 调整窗位、窗宽
img_data = pixel_array
winwidth = 500
wincenter = 50
rows = dcm.Rows
cols = dcm.Columns
dcm_temp = setDicomWinWidthWinCenter(img_data, winwidth, wincenter, rows, cols)
# 可视化
dcm_img = Image.fromarray(dcm_temp) # 将Numpy转换为PIL.Image
dcm_img = dcm_img.convert('L')
# plt.imshow(img, cmap=plt.cm.bone)
# 保存为jpg文件,用作后面的生成label用
dcm_img.save('../output/temp.jpg')
# 显示图像
dcm_img.show()同时读取一个文件夹中的 dicom 文件,并处理保存 (写成循环即可)
def main_mulit(path):
names = os.listdir(path) # 读取文件夹中的所有文件名
for i in range(len(names)):
dicom_name = path+names[i]
dcm = pydicom.read_file(dicom_name) # 读取 dicom 文件
pixel_array, dcm.Rows, dcm.Columns = get_pixeldata(dcm) # 得到 dicom文件的 CT 值
img_data = pixel_array
winwidth = 500
wincenter = 50
rows = dcm.Rows
cols = dcm.Columns
dcm_temp = setDicomWinWidthWinCenter(img_data, winwidth, wincenter, rows, cols) # 调整窗位、窗宽
# 可视化
dcm_img = Image.fromarray(dcm_temp) # 将 Numpy转换为 PIL.Image
dcm_img = dcm_img.convert('L')
# 批量保存
dcm_img.save("C:/output/%s_%s.png" % (path1, names[i]))
注意,以上都写成函数的形式,运行时需要调用,并注意文件路径的修改。
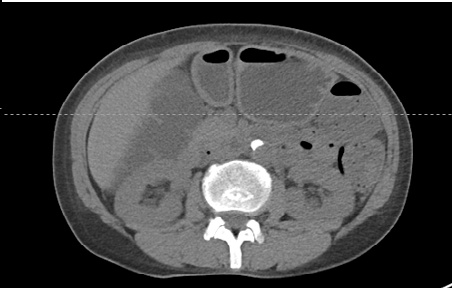
单张dicom文件处理
main_single()
批量处理
main_mulit(path)
公众号文章链接:
图像识别 | 使用Python对医学Dicom文件的预处理(含代码)mp.weixin.qq.com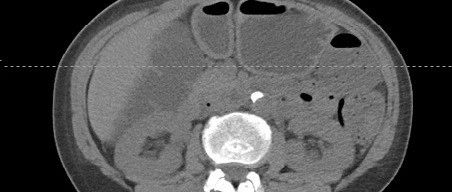
觉得有收获的话,麻烦点个赞再走呗~
如果觉得我的文章对您有用,请随意打赏。你的支持将鼓励我继续创作!
